setup
i came across a spreadsheet of the 2023 version of Journal Impact Factor (JIF). a quick look in Excel showed that the JIFs appeared to decrease from 500+ to a long stretch of 1.6s that seemed to go on forever. so i'll need to check if this spreadsheet's complete or correct, and if both yes, i'll make a suitable copy of it for picking candidate journals for reserach articles.
solution
the plan
- load the JIF spreadsheet in R.
- check there's something obviously wrong with the JIFs and number of journals.
- if the source table seems trustworthy enough, then a. pick journal by field. fields w/ a shred of possibility of publishing something biomedical or clinical are included. b. sort journals by descending JIF.
- write the resulting simpler JIF table to a new sheet in the source doc.
programming language & module(s):
- R
- tidyverse
- readxl
- openxlsx
input docs
- can be found in here: https://discuss.sci-hub.org.cn/d/2605
variables to customize:
folderandjif_name, for determining JIF spreadsheet doc path
the script:
sort_out_JIFs.R
| rm(list = ls()) | |
| for (pkg in c('tidyverse', 'readxl', 'openxlsx')){ | |
| if(!requireNamespace(pkg, quietly = T)){ | |
| install.packages(pkg, dependencies = T) | |
| } | |
| library(pkg, character.only = T) | |
| } | |
| folder <- 'c:/users/xiao/desktop' | |
| jif_name <- 'JIF.2024.xlsx' | |
| jif_path <- file.path(folder, jif_name) | |
| df_input <- read_excel(jif_path) | |
| df_input |> glimpse | |
| # 21800+ journals are included in the 2024 JCR according to official website | |
| # (see https://clarivate.com.cn/2024/06/20/2024jcr/). so the total journal number | |
| # matches. | |
| df_input |> | |
| count(JIF) |> | |
| arrange(desc(JIF)) |> | |
| print(n = df_input |> | |
| nrow) | |
| # 13 JIF == 'N/A's and 683 '<0.1's. | |
| print(683 + 13) | |
| df_input <- df_input |> | |
| select(c(1, 2, 5, 6, 7)) |> | |
| mutate(JIF = JIF |> as.numeric) | |
| df_input |> head | |
| df_input |> | |
| count(JIF) |> | |
| arrange(desc(JIF)) |> | |
| print(n = df_input |> nrow) | |
| # so the 696 NAs resulted form the <0.1 and N/A. | |
| df_input <- df_input |> | |
| mutate( | |
| Category_level_1 = Category |> | |
| str_split('\\|') |> | |
| sapply(function(x) x[[1]])) | |
| # pick fields to keep in the simplified JIF table. | |
| field_level_1_full <- df_input |> | |
| distinct(Category_level_1) |> | |
| pull(Category_level_1) |> | |
| sort | |
| field_level_1_full | |
| field_level_1_keep <- field_level_1_full[ | |
| c( | |
| 2,3,4,5,7,8,9,1011,12,14,16,20:26,29:38,40,51,52,55,56,57,58,59,60,65,66, | |
| 67,83,89,94:97,103,104,106,108,115,116,118,121,138,139,152:157,161,162, | |
| 165,167:170,172:174,176:178,180,181,184:188,198,199,207,219,224,225,226, | |
| 239,243,247,249,250,251,254 | |
| ) | |
| ] | |
| field_level_1_keep |> head | |
| df_jif <- df_input |> | |
| filter(Category_level_1 %in% field_level_1_keep) |> | |
| mutate( | |
| # make Category_level_1 more readable | |
| Category_level_1 = Category_level_1 |> str_to_lower, | |
| # convert to numbers in case i'd want to sort by this col in excel | |
| JIF5Years = JIF5Years |> as.numeric | |
| ) |> | |
| arrange(desc(JIF), Name) | |
| df_jif |> head | |
| # looking good. | |
| # export to a new sheet in the spreadsheet src doc. | |
| jif_xlsx <- loadWorkbook(jif_path) | |
| out_sheet_name <- 'JIF_processed' | |
| if (out_sheet_name %in% names(jif_xlsx)){ | |
| jif_xlsx |> removeWorksheet(out_sheet_name) | |
| } | |
| jif_xlsx |> addWorksheet(out_sheet_name) | |
| jif_xlsx |> writeData(sheet = out_sheet_name, x = df_jif) | |
| # set xlsx doc font while i'm at it ... | |
| jif_xlsx |> modifyBaseFont( | |
| fontSize = 11, | |
| fontColour = 'black', | |
| fontName = 'Times New Roman') | |
| jif_xlsx |> saveWorkbook(jif_path, overwrite = T) |
output
the simplied JIF table can be found in the sheet title 'JIF_processed' in the source JIF spreadsheet doc. it should look like this:
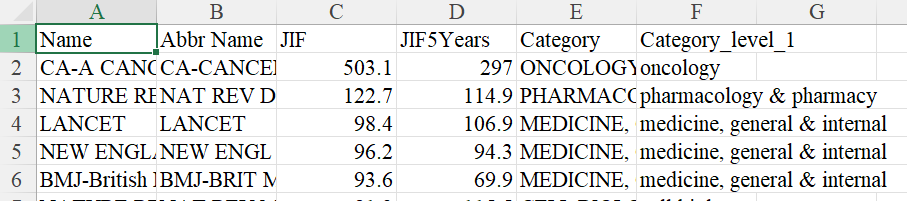
note to self
- the
applyfunction family's super handy. always think of them when feeiling an itch for theforloop. - there may be somthing odd about using
|in regex, since'a|b' |> str_split('|')returns a weird list of[[1]] [1] "" "a" "|" "b" ""instead of[[1]] [1] "a" "b". for the latter, usestr_split('\\|').